Fazhan Shi et al., proposed and applied quantum technology to detect electron spin resonance spectra of single DNA in aqueous solutions at room temperature for the first time. It represents an important step toward single molecule magnetic resonance investigation of biomolecules in their native environments. The research results have been published in Nature Methods [Nature Methods 15, 697–699 (2018)].

Figure | Detection of single DNA molecule in aqueous solution with diamond sensor.
Magnetic resonance is essential in revealing the structure and dynamics of biomolecules. However, traditional magnetic resonance technology is limited by detection sensitivity, and cannot realize single-molecule research. Its research object is usually a multi-billion-molecule macroscopic system. Our group employing NV center in diamond as a highly sensitive quantum sensor, which forms a quantum interferometer under the modulation of green laser and specific frequency microwave pulses. The weak magnetic signals can be transform into a quantum phase signal and read out optically. At the same time, due to the size of the diamond sensor at the atomic level, nanoscale spatial resolution can be achieved. Therefore, the use of NV centers is a promising avenue for single molecule detection.
Our group previously reported single-molecule magnetic resonance spectroscopy of proteins [Science 347, 1135–1138 (2015)], and achieved a technological breakthrough in single-molecule magnetic resonance, in which the target were fixed on the diamond surface by biological glue. However, the majority of biomolecules function in aqueous solution under ambient temperature, where they undergo a range of motions. The single-molecule magnetic resonance detection in native-like environment is the only way to study its biological functions. Our group, collaborated with Prof. Peter Z. Qin of the University of Southern California, used NV centers to detect ESR spectra of individual, tethered DNA duplexes in aqueous buffer solutions at ambient temperatures.
First, in order to prevent the diffusion of DNA molecules in solution, we adapted a chemical reaction process in which one strand of the DNA molecule (indicated by the red dotted line in the figure below) was attached to the diamond surface, then hybridized with a complementary strand with a spectroscopic label. It ensures the uniform distribution of DNA molecules on the diamond surface. Secondly, we implemented a diamond pillar array design, and improved microwave control technology which reduced the detection time from hours to minutes. In the end, we successfully obtained the magnetic resonance spectrum of a single DNA molecule in an aqueous environment, and obtained its dynamic and environmental characteristics by spectral analysis. A rotational correlation time of the DNA molecule is obtained by spectral broadening and the local hydrophobic environment information is obtained by the hyperfine couplings.
This work provides a new method for studying the structure and function of individual biomolecules in an aqueous environment and is an important step toward single molecule studies of cells in situ. Combined with scanning probes, gradient magnetic fields and other technologies, the technology can be applied to single-molecule imaging, structural analysis and dynamics detection in the life science field in the future and understand biological characteristics and life functions from a single molecule level. Reviewers commented on the work: “Single molecule techniques are crucial for modern life sciences and demonstration single DNA detection and observation of DNA dynamics will be of big interest for wide community of scientists.” The work was published in Nature Methods and was selected as one of the five cover titles.
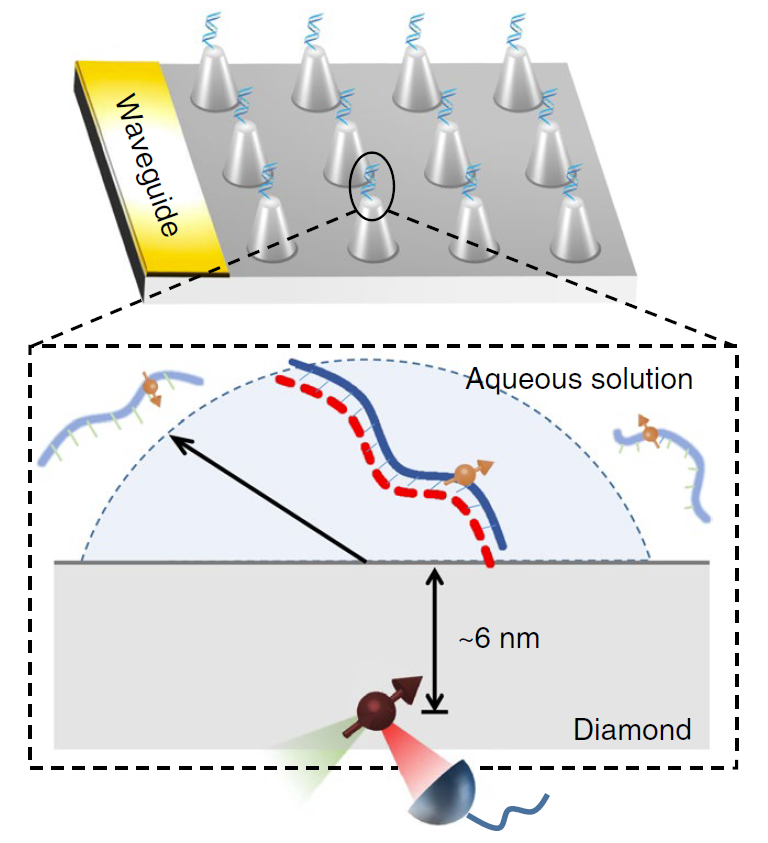
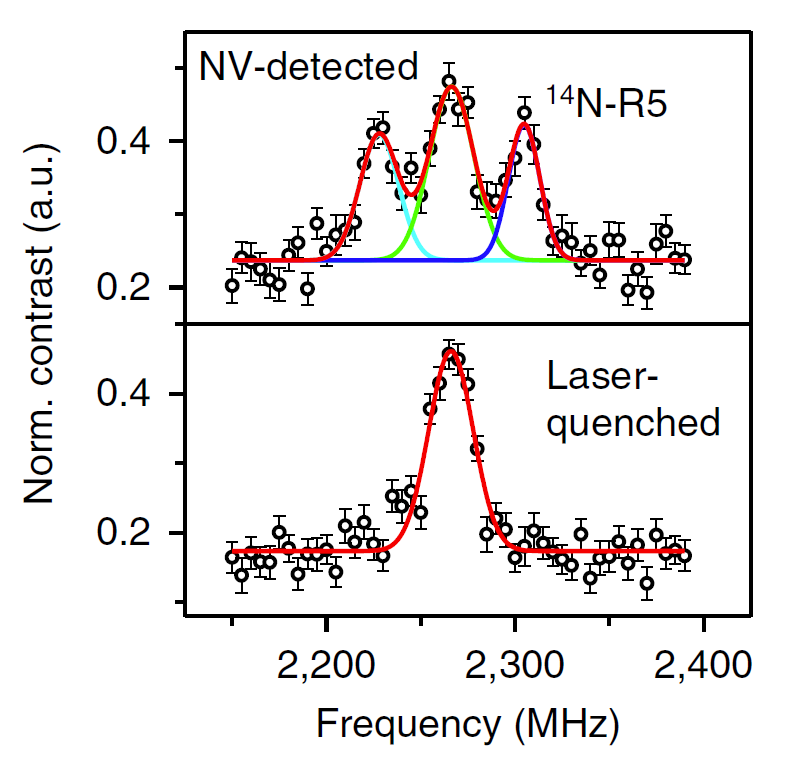
FIGURE left, Experimental scheme. right, NV-detected spectrum of a single 14N-spin-labeled DNA